Commits on Source (184)
Showing
- .gitlab-ci.yml 30 additions, 2 deletions.gitlab-ci.yml
- 0-collect-data/datacite-dois.txt 1644 additions, 1181 deletions0-collect-data/datacite-dois.txt
- 0-collect-data/datacite.py 2 additions, 15 deletions0-collect-data/datacite.py
- 0-collect-data/nakala-uga-users.txt 15 additions, 1 deletion0-collect-data/nakala-uga-users.txt
- 0-collect-data/rdg.py 7 additions, 7 deletions0-collect-data/rdg.py
- 0-collect-data/zenodo-dois.txt 724 additions, 995 deletions0-collect-data/zenodo-dois.txt
- 0-collect-data/zenodo.py 11 additions, 15 deletions0-collect-data/zenodo.py
- 1-enrich-with-datacite/all_datacite_clients_for_uga.csv 44 additions, 0 deletions1-enrich-with-datacite/all_datacite_clients_for_uga.csv
- 1-enrich-with-datacite/concatenate-enrich-dois.py 64 additions, 3 deletions1-enrich-with-datacite/concatenate-enrich-dois.py
- 1-enrich-with-datacite/nb-dois.txt 1 addition, 0 deletions1-enrich-with-datacite/nb-dois.txt
- 1-enrich-with-datacite/z_personal_functions.py 31 additions, 2 deletions1-enrich-with-datacite/z_personal_functions.py
- 2-produce-graph/all_datacite_clients.csv 0 additions, 35 deletions2-produce-graph/all_datacite_clients.csv
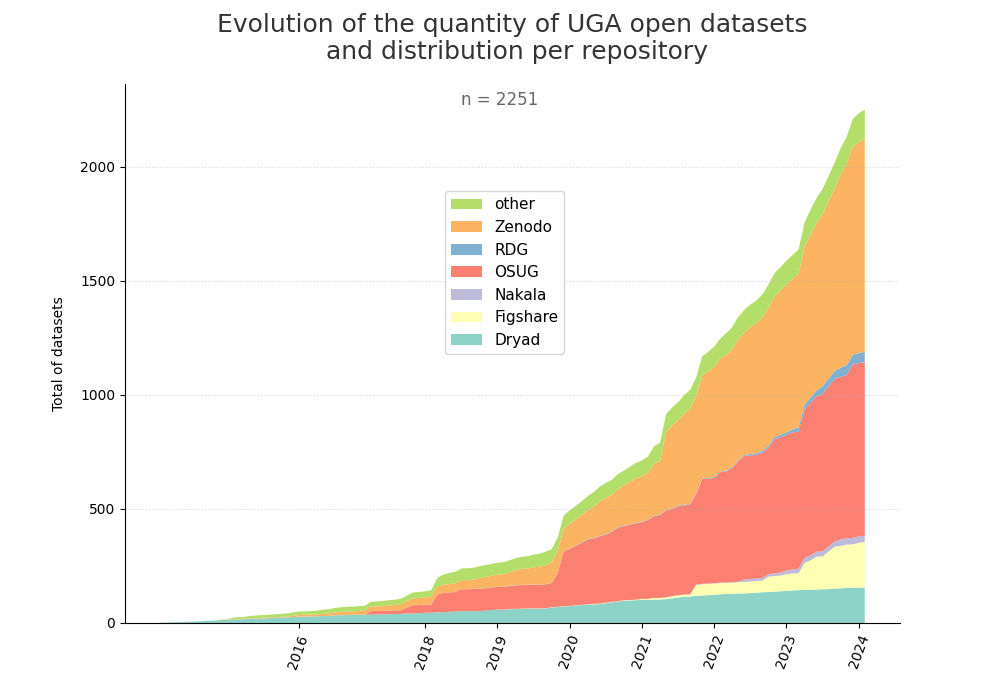
- 2-produce-graph/hist-evol-datasets-per-repo.png 0 additions, 0 deletions2-produce-graph/hist-evol-datasets-per-repo.png
- 2-produce-graph/hist-evol-datasets-per-repo.py 2 additions, 3 deletions2-produce-graph/hist-evol-datasets-per-repo.py
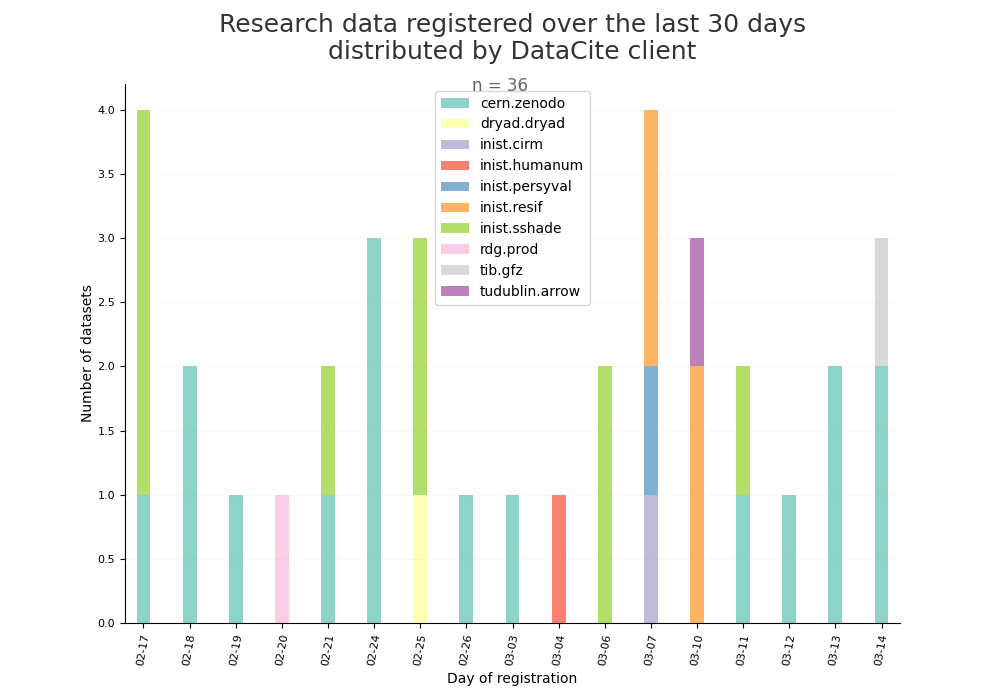
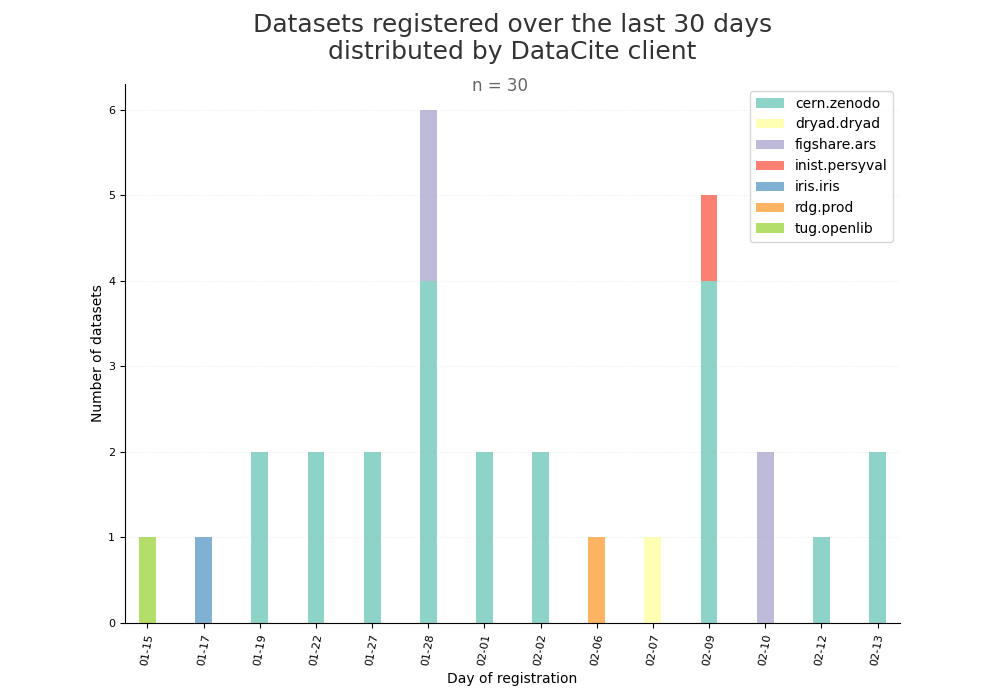
- 2-produce-graph/hist-last-datasets-by-client.png 0 additions, 0 deletions2-produce-graph/hist-last-datasets-by-client.png
- 2-produce-graph/hist-last-datasets-by-client.py 1 addition, 1 deletion2-produce-graph/hist-last-datasets-by-client.py
- 2-produce-graph/hist-quantity-year-type.png 0 additions, 0 deletions2-produce-graph/hist-quantity-year-type.png
- 2-produce-graph/hist-quantity-year-type.py 2 additions, 2 deletions2-produce-graph/hist-quantity-year-type.py
- 2-produce-graph/pie--datacite-client.png 0 additions, 0 deletions2-produce-graph/pie--datacite-client.png
- 2-produce-graph/pie--datacite-type.png 0 additions, 0 deletions2-produce-graph/pie--datacite-type.png
This diff is collapsed.
This diff is collapsed.
1-enrich-with-datacite/nb-dois.txt
0 → 100644
This diff is collapsed.
This diff is collapsed.
| W: | H:
| W: | H:


This diff is collapsed.
| W: | H:
| W: | H:


This diff is collapsed.
This diff is collapsed.
This diff is collapsed.
This diff is collapsed.
This diff is collapsed.