Execution du pipeline. Actualisation des dois et des graphes.
Showing
- 1-enrich-with-datacite/all_datacite_clients_for_uga.csv 2 additions, 2 deletions1-enrich-with-datacite/all_datacite_clients_for_uga.csv
- 1-enrich-with-datacite/nb-dois.txt 1 addition, 1 deletion1-enrich-with-datacite/nb-dois.txt
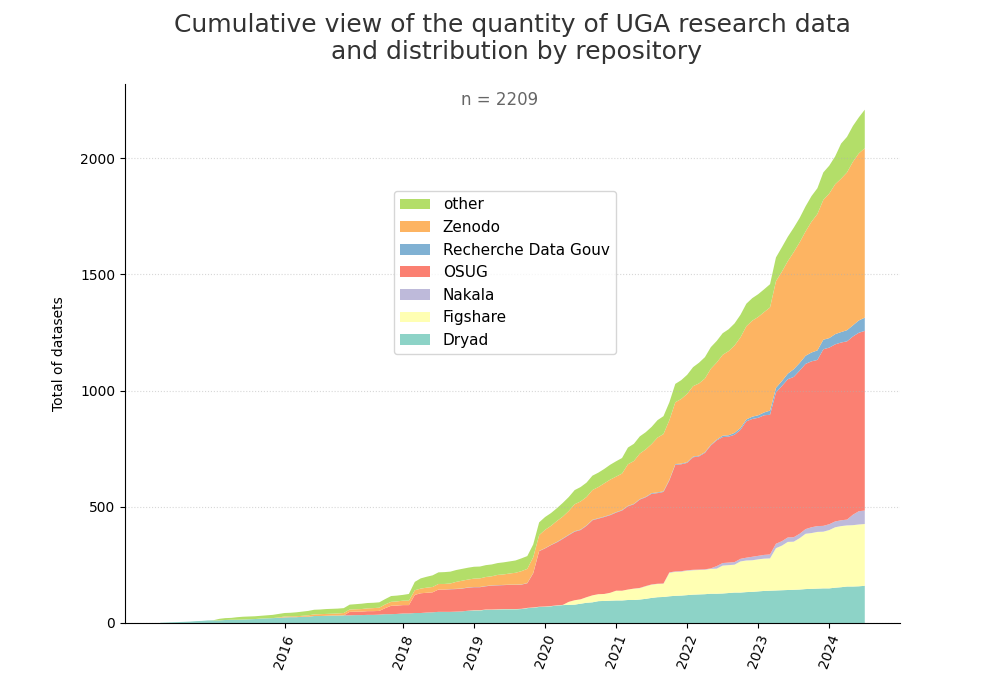
- 2-produce-graph/hist-evol-datasets-per-repo.png 0 additions, 0 deletions2-produce-graph/hist-evol-datasets-per-repo.png
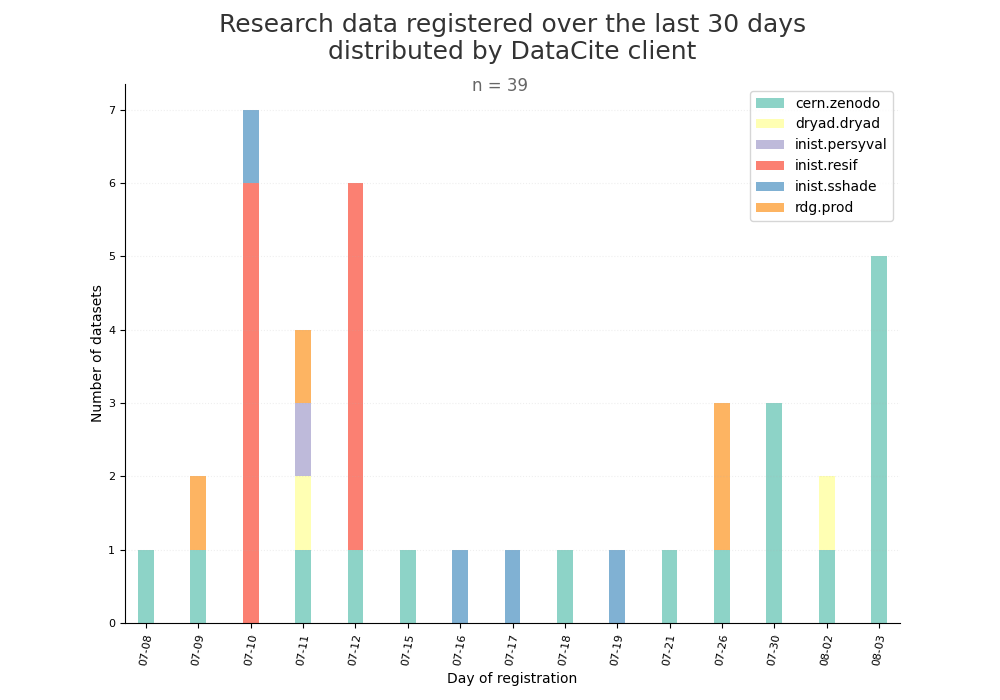
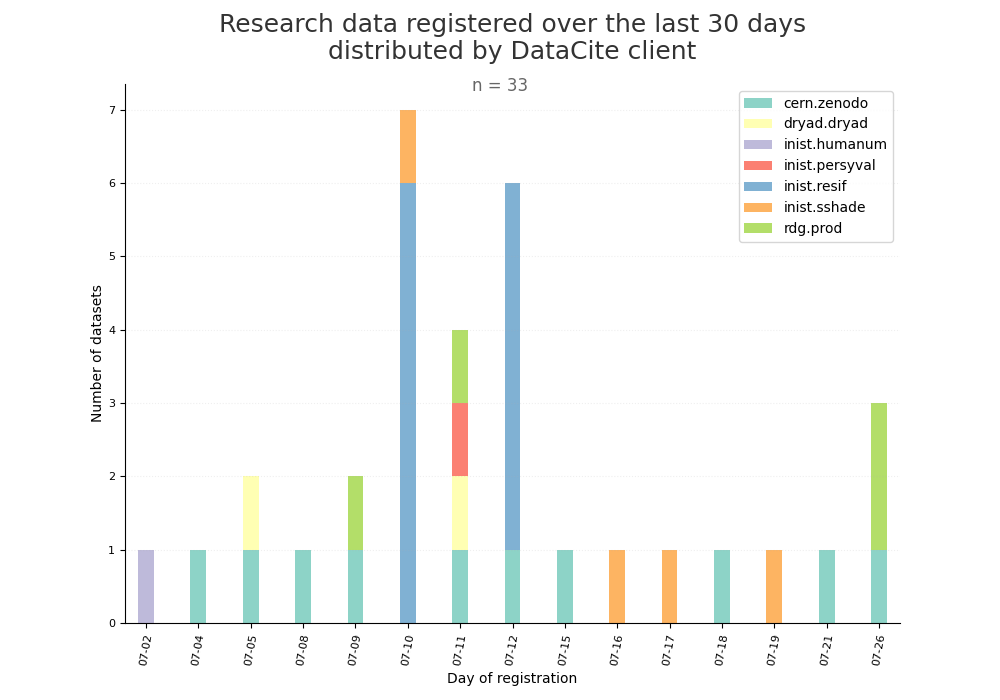
- 2-produce-graph/hist-last-datasets-by-client.png 0 additions, 0 deletions2-produce-graph/hist-last-datasets-by-client.png
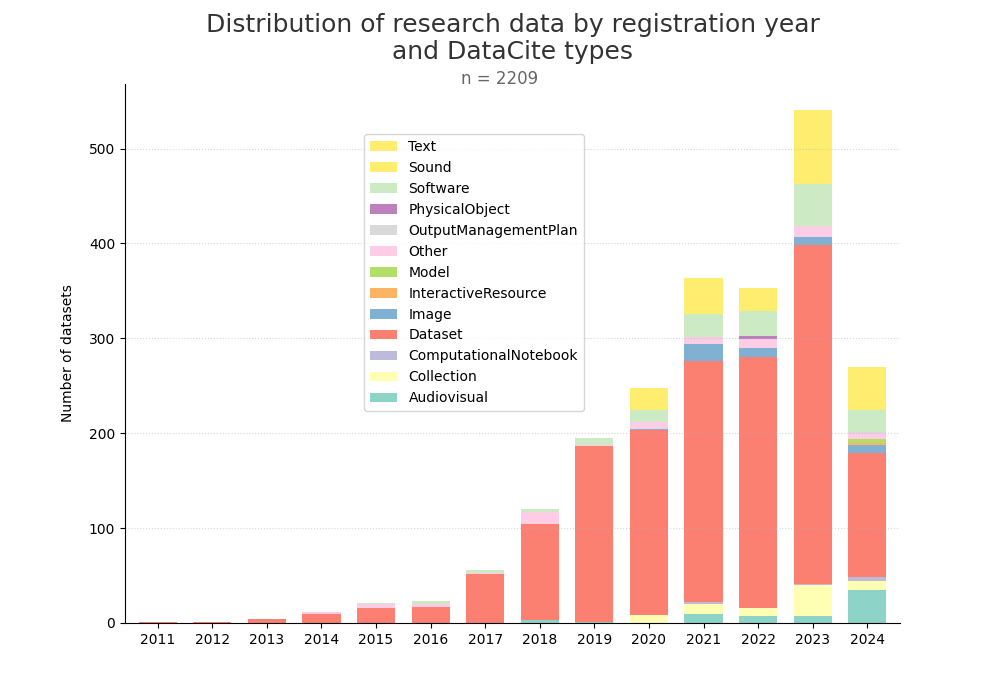
- 2-produce-graph/hist-quantity-year-type.png 0 additions, 0 deletions2-produce-graph/hist-quantity-year-type.png
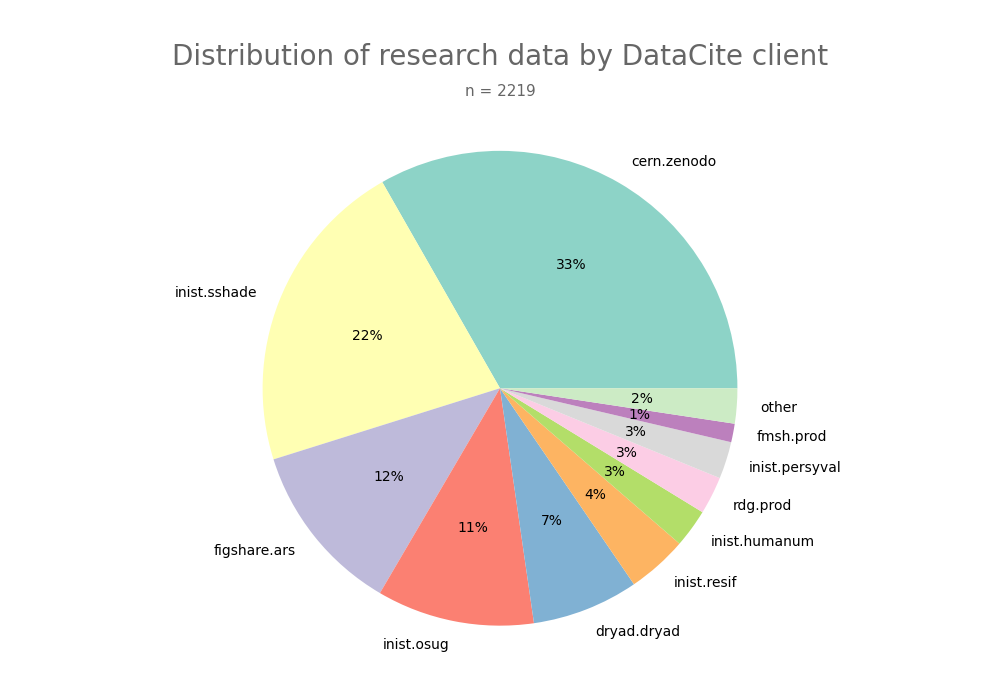
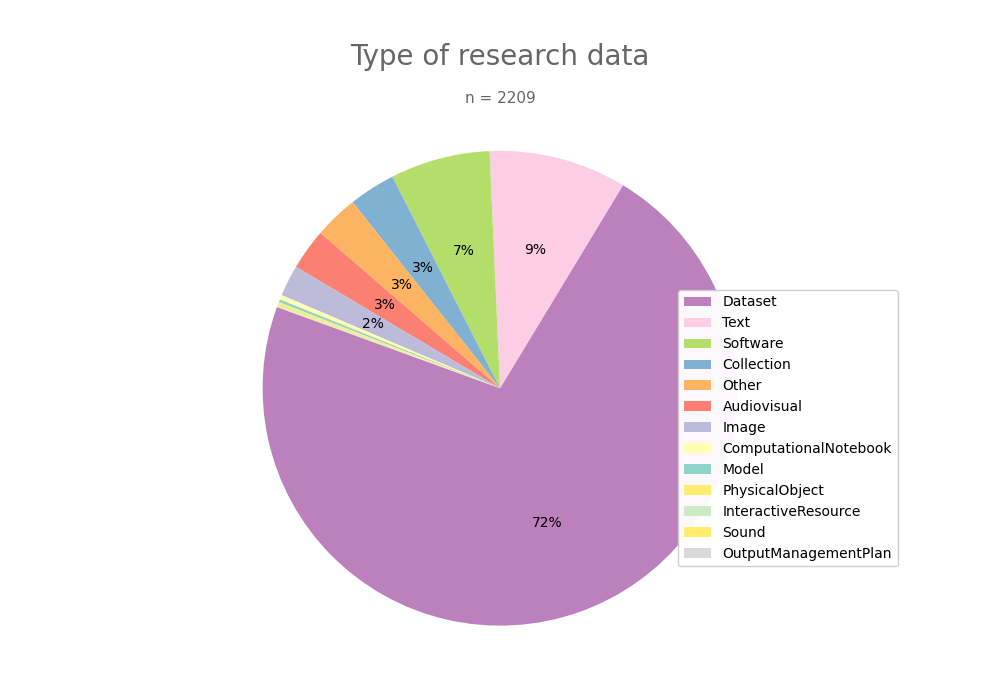
- 2-produce-graph/pie--datacite-client.png 0 additions, 0 deletions2-produce-graph/pie--datacite-client.png
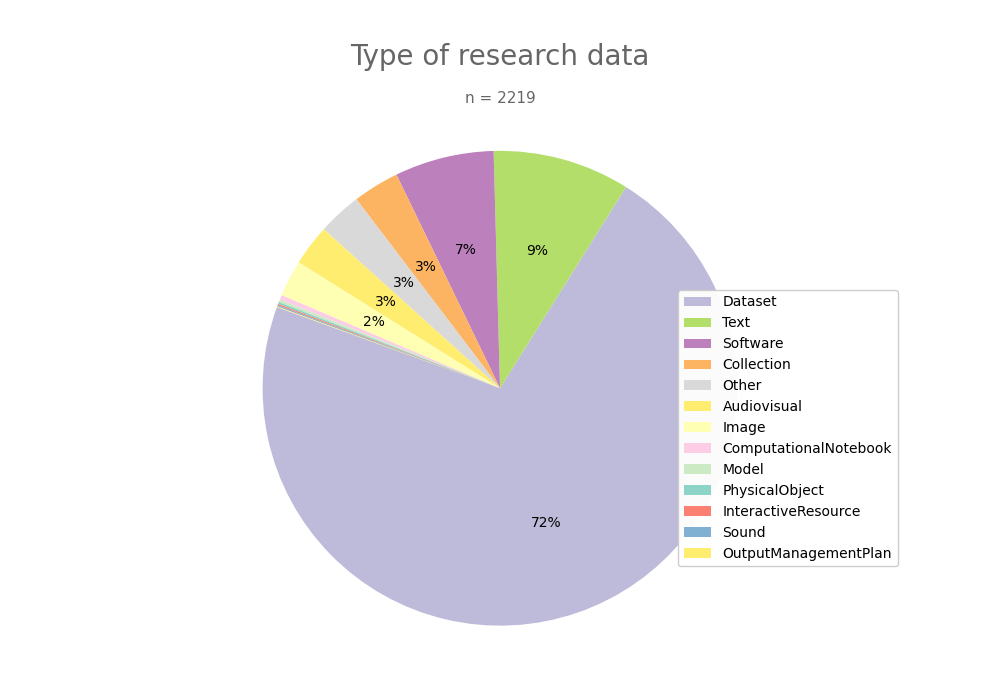
- 2-produce-graph/pie--datacite-type.png 0 additions, 0 deletions2-produce-graph/pie--datacite-type.png
- dois-uga.csv 30 additions, 0 deletionsdois-uga.csv
| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:



| W: | H:
| W: | H:


| W: | H:
| W: | H: