add step 1 and 2
Showing
- .gitignore 1 addition, 1 deletion.gitignore
- 1-enrich-with-datacite/concatenet-enrich-dois.py 60 additions, 0 deletions1-enrich-with-datacite/concatenet-enrich-dois.py
- 1-enrich-with-datacite/datacite-parser-instructions.json 66 additions, 0 deletions1-enrich-with-datacite/datacite-parser-instructions.json
- 1-enrich-with-datacite/z_personal_functions.py 192 additions, 0 deletions1-enrich-with-datacite/z_personal_functions.py
- 2-produce-graph/hist--datasets-by-year.png 0 additions, 0 deletions2-produce-graph/hist--datasets-by-year.png
- 2-produce-graph/hist-quantity-by-repo.py 39 additions, 0 deletions2-produce-graph/hist-quantity-by-repo.py
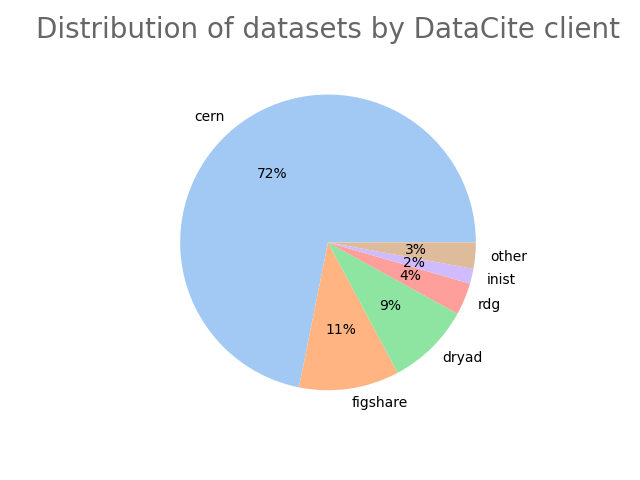
- 2-produce-graph/pie--datacite-client.png 0 additions, 0 deletions2-produce-graph/pie--datacite-client.png
- 2-produce-graph/pie-datacite-client.py 30 additions, 0 deletions2-produce-graph/pie-datacite-client.py
- 2-produce-graph/z_my_functions.py 14 additions, 0 deletions2-produce-graph/z_my_functions.py
- README.md 9 additions, 1 deletionREADME.md
- dois-uga.csv 2827 additions, 0 deletionsdois-uga.csv
- run-all-codes.py 1 addition, 2 deletionsrun-all-codes.py
2-produce-graph/hist--datasets-by-year.png
0 → 100644
25.6 KiB
2-produce-graph/hist-quantity-by-repo.py
0 → 100644
2-produce-graph/pie--datacite-client.png
0 → 100644
30 KiB
2-produce-graph/pie-datacite-client.py
0 → 100644
2-produce-graph/z_my_functions.py
0 → 100644
dois-uga.csv
0 → 100644
This diff is collapsed.