Commits on Source (86)
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored7c03fc71
-
Elias Chetouane authored0d7e31cc
-
Elias Chetouane authoredf983fe44
-
Elias Chetouane authored0f1c52ce
-
Elias Chetouane authored60e83e2e
-
Maxence Larrieu authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authorede8b7601a
-
Elias Chetouane authored
-
Elias Chetouane authored16e22d91
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authorede4cd98ec
-
Elias Chetouane authorede2e466d5
-
Elias Chetouane authoredd87f6251
-
Elias Chetouane authoredfd036b3c
-
Elias Chetouane authoredb3c8eea6
-
Maxence Larrieu authored9384e0b2
-
Maxence Larrieu authored
-
Elias Chetouane authored708ac45f
-
Elias Chetouane authoredb78accb8
-
Elias Chetouane authoreda2116550
-
Elias Chetouane authoreddd409503
-
Elias Chetouane authored
Prise en compte des relations des jeux de données, versions, jeux identiques... See merge request !3
-
Elias Chetouane authored9751c5f8
-
Maxence Larrieu authored45637a13
-
Elias Chetouane authored44279d03
-
Elias Chetouane authored
-
Elias Chetouane authoreda62d8cd2
-
Elias Chetouane authored3f153593
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authoredfa5017a9
-
Maxence Larrieu authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authoredb876db69
-
Maxence Larrieu authored
-
Elias Chetouane authored559a5f18
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored9ef67938
-
Maxence Larrieu authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored512cc0a9
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored91441863
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authoreda36ebdb7
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored
-
Elias Chetouane authored2bd53d86
Showing
- .gitlab-ci.yml 2 additions, 1 deletion.gitlab-ci.yml
- 0-collect-data/nakala-uga-users.txt 15 additions, 1 deletion0-collect-data/nakala-uga-users.txt
- 0-collect-data/rdg.py 6 additions, 6 deletions0-collect-data/rdg.py
- 1-enrich-with-datacite/all_datacite_clients_for_uga.csv 36 additions, 26 deletions1-enrich-with-datacite/all_datacite_clients_for_uga.csv
- 1-enrich-with-datacite/concatenate-enrich-dois.py 27 additions, 0 deletions1-enrich-with-datacite/concatenate-enrich-dois.py
- 1-enrich-with-datacite/nb-dois.txt 1 addition, 1 deletion1-enrich-with-datacite/nb-dois.txt
- 1-enrich-with-datacite/z_personal_functions.py 25 additions, 9 deletions1-enrich-with-datacite/z_personal_functions.py
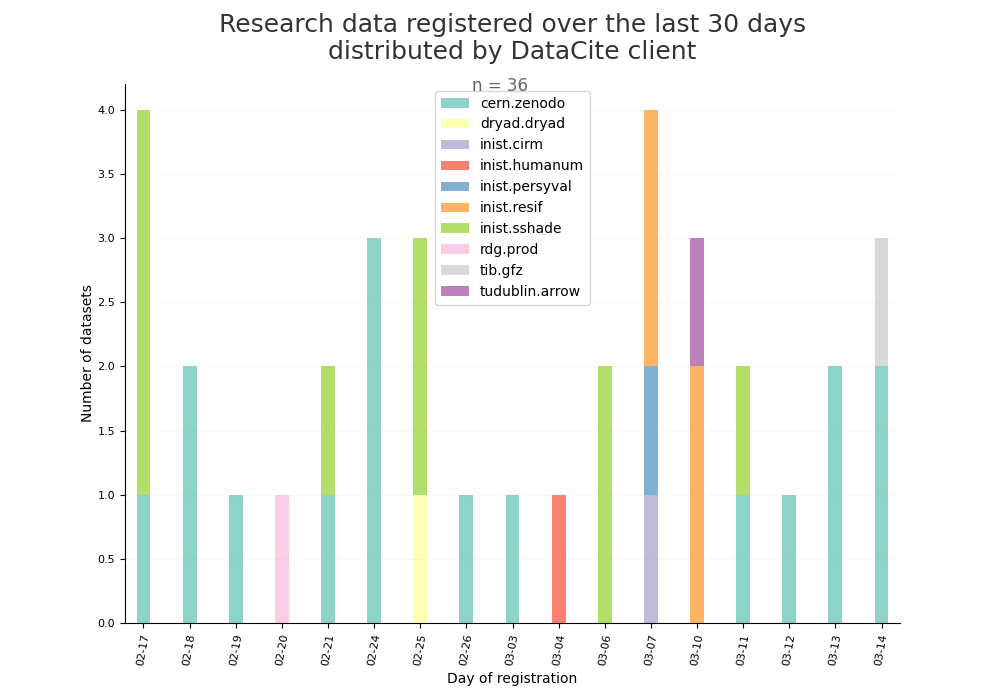
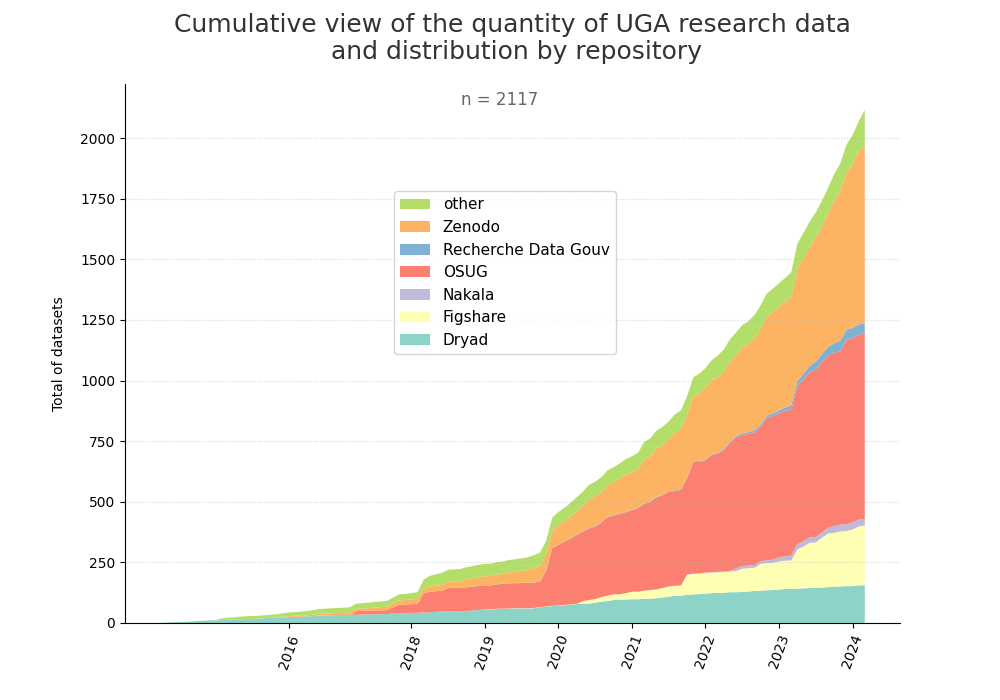
- 2-produce-graph/hist-evol-datasets-per-repo.png 0 additions, 0 deletions2-produce-graph/hist-evol-datasets-per-repo.png
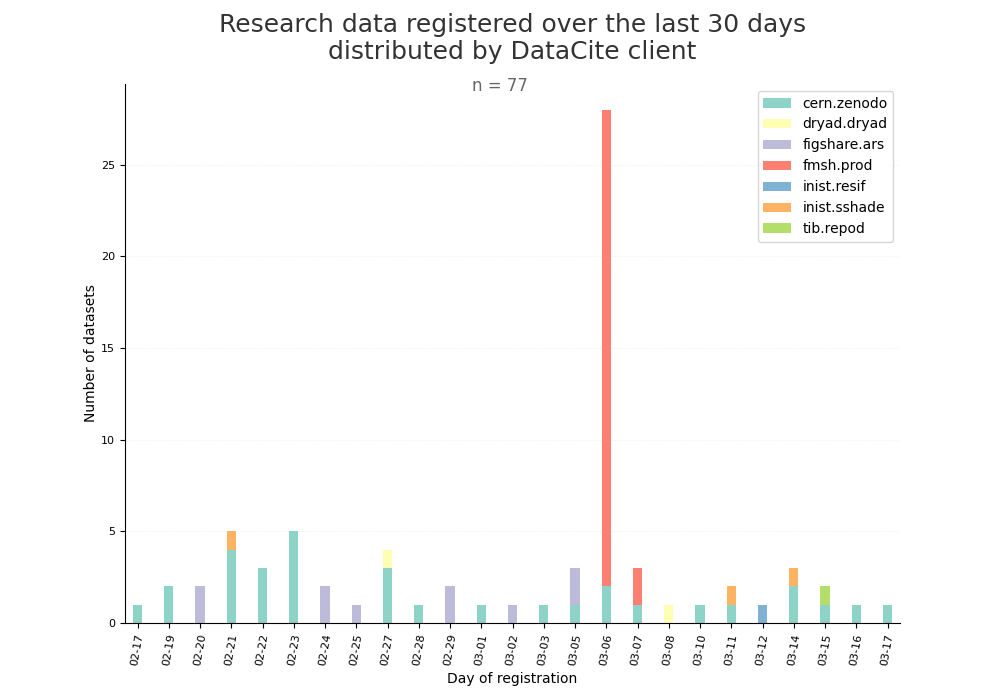
- 2-produce-graph/hist-last-datasets-by-client.png 0 additions, 0 deletions2-produce-graph/hist-last-datasets-by-client.png
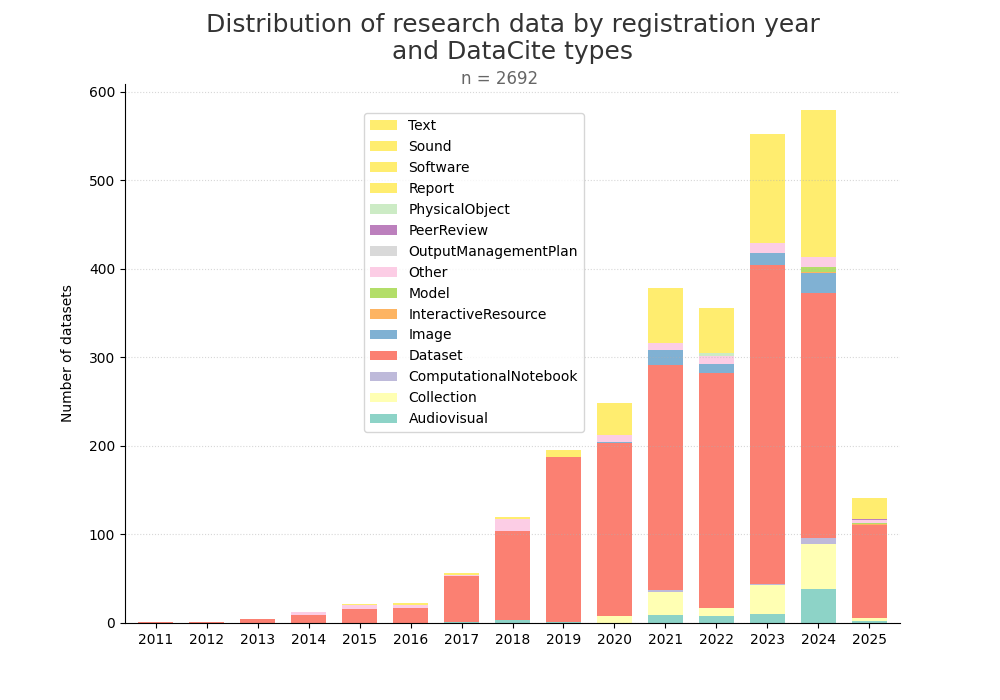
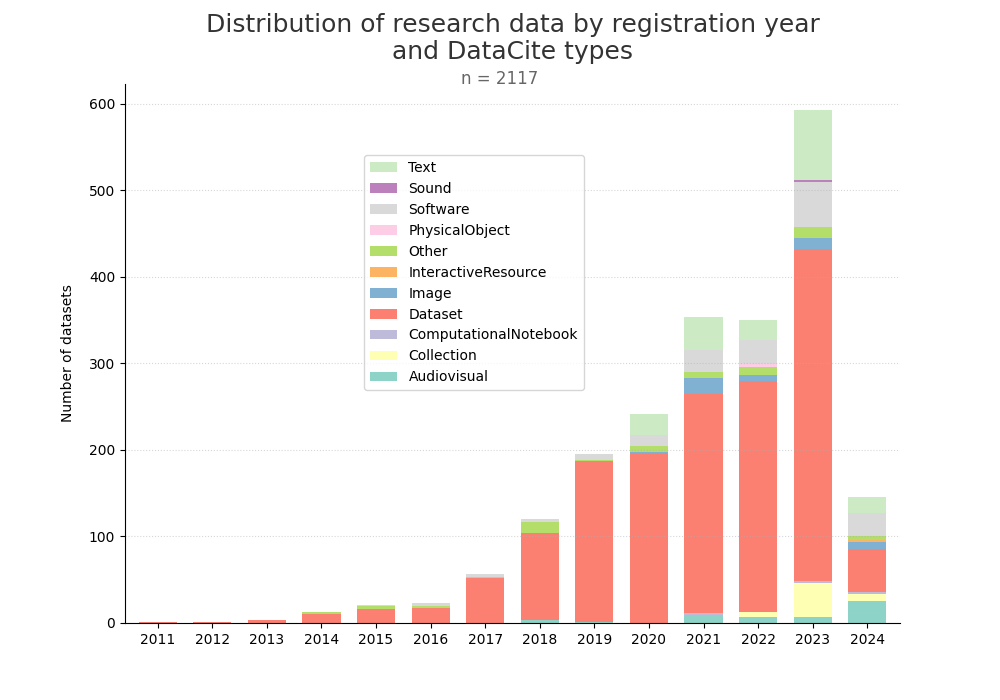
- 2-produce-graph/hist-quantity-year-type.png 0 additions, 0 deletions2-produce-graph/hist-quantity-year-type.png
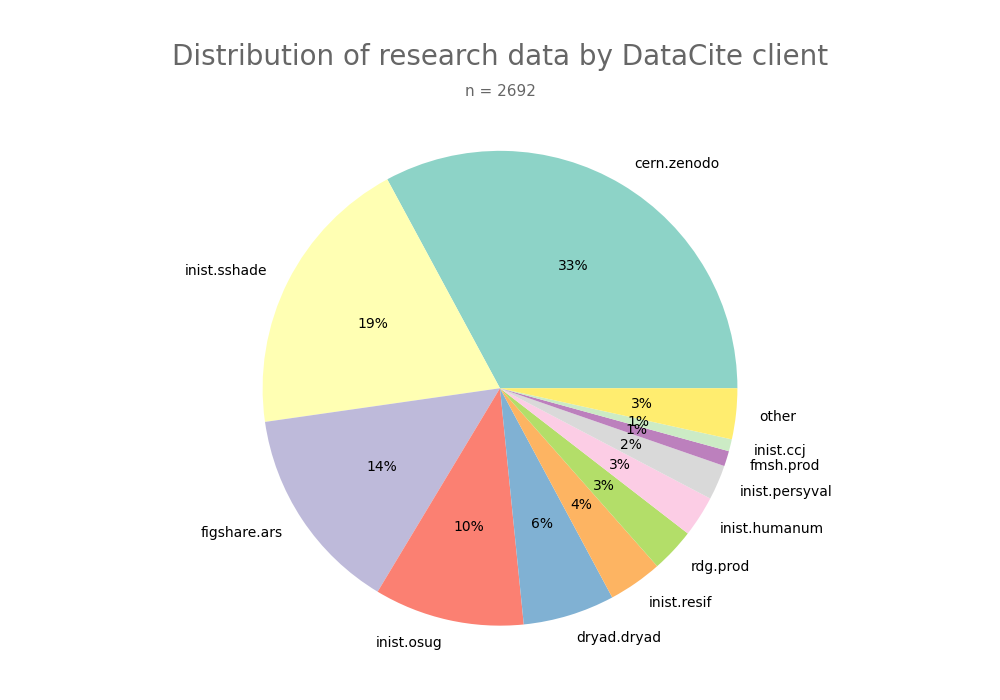
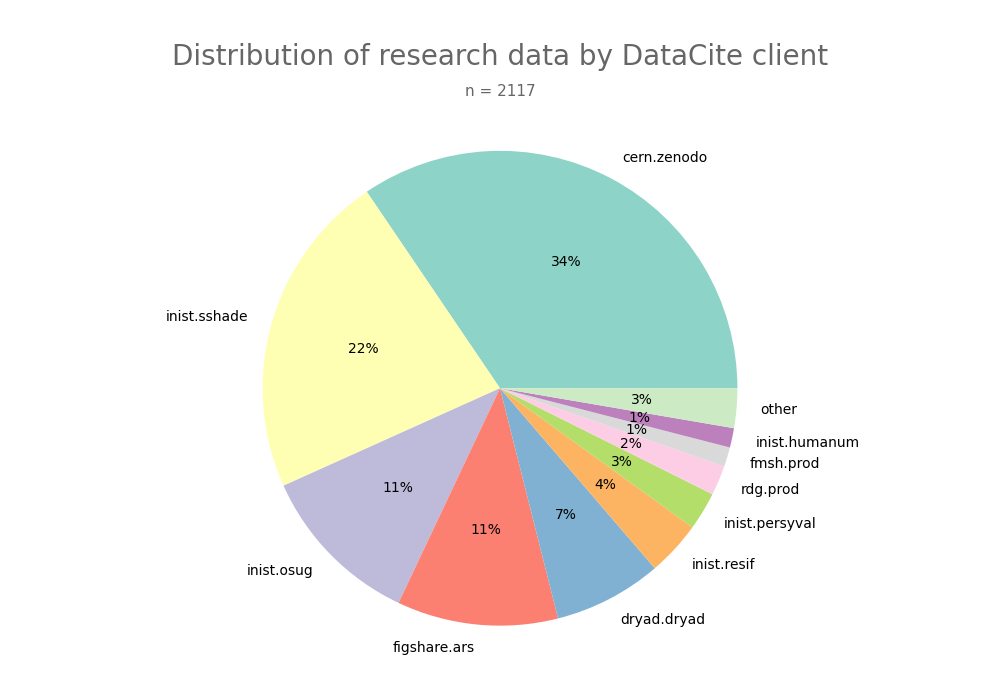
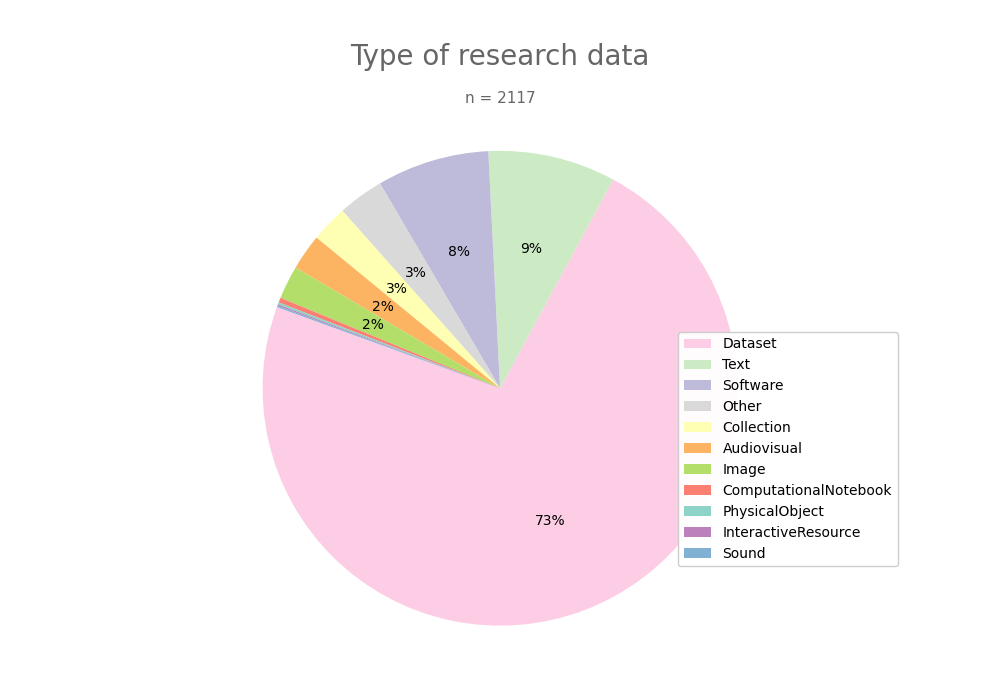
- 2-produce-graph/pie--datacite-client.png 0 additions, 0 deletions2-produce-graph/pie--datacite-client.png
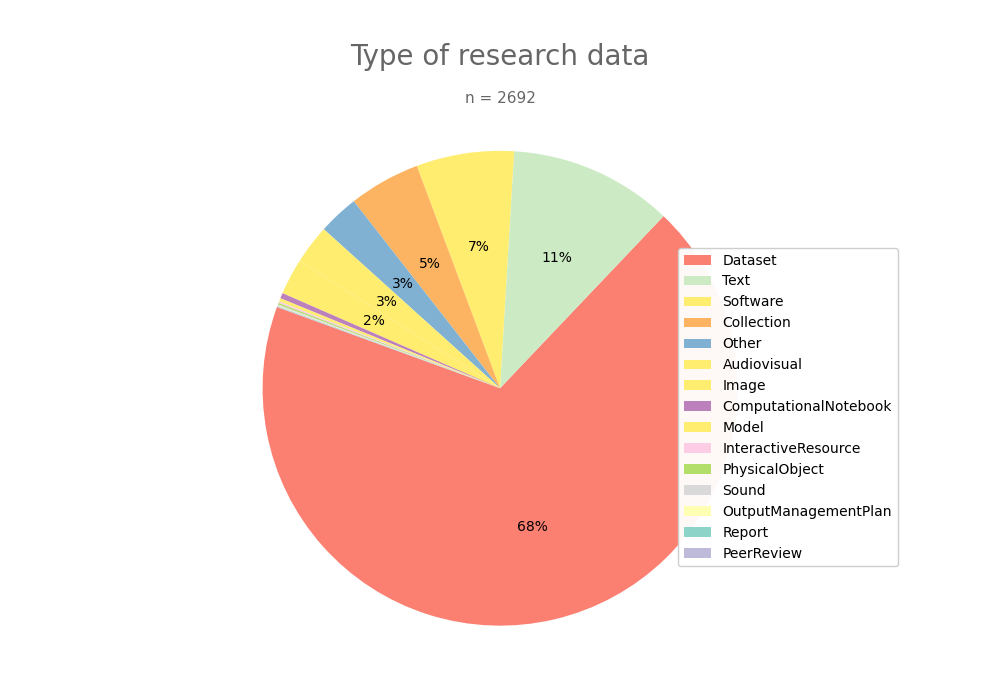
- 2-produce-graph/pie--datacite-type.png 0 additions, 0 deletions2-produce-graph/pie--datacite-type.png
- README.md 22 additions, 7 deletionsREADME.md
- dois-uga--last-500.csv 538 additions, 0 deletionsdois-uga--last-500.csv
- dois-uga.csv 12023 additions, 4602 deletionsdois-uga.csv
- notes-feedback.md 0 additions, 2 deletionsnotes-feedback.md
| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:


dois-uga--last-500.csv
0 → 100644
source diff could not be displayed: it is too large. Options to address this: view the blob.